Postdoc Research Spotlight: College of Agriculture & Life Sciences
Highlighting Postdoc Research from the College Agriculture & Life Sciences
 Javier Brumos, Ph.D.
Javier Brumos, Ph.D.
Postdoc in Plant & Microbial Biology
Understanding the regulation of genes associated with fruit ripening to reduce spoilage
Research Blurb:
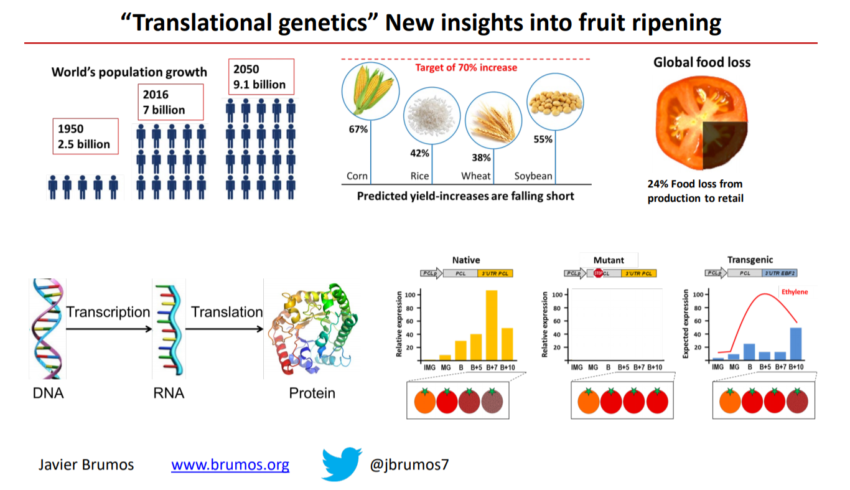
One of the major challenges of modern agriculture is to reduce crop losses which are mainly caused by over-ripening and senescence. A better understanding of how the ripening process is regulated could result in direct applications to reduce spoilage. The plant hormone ethylene regulates the ripening process and gene expression changes in response to this hormone during fruit ripening have been extensively studied at the transcriptional level, but little is known about ripening-associated shifts in the efficiency of transcript translation.
Our laboratory has pioneered the implementation of the ribosome footprinting technology (Ribo-seq) in plants and currently we are using it to monitor changes in transcript translation at a whole-genome scale and single-codon resolution on developing fruits. Ribo-seq and RNA-seq will aid to identify transcripts that change their translation efficiency during the ripening process. This approach will shed light on the molecular basis of translational regulation and, perhaps, unravel new ways to control gene activity in agriculturally important processes, such as fruit ripening, and could contribute to the development of new crop varieties as well as lead to improved agricultural practices.
Key Finding:
In parallel, we have characterized regulatory elements that are required and sufficient for the translational inhibition of gene expression in the presence of the endogenous ripening hormone ethylene. We are testing their ability to delay fruit softening and control fruit texture without compromising other ripening associated traits, such as flavor, color, and nutritional value.
Delve Deeper into Javier’s Publications
Research Impact:
Agricultural production can be increased by improving germplasm quality and yield, both of which are expected to continue to grow, but at a slower pace than in past decades. Therefore, alternative/complementary approaches are needed to boost agricultural production without exploiting nonrenewable resources, or increasing the use of arable land, energy, and water. Consequently, to achieve the desired food-availability goals, it is essential to develop sustainable means of increasing food production, as well as to minimize food waste.
This project is the foundation for future in-depth studies of novel translation regulation mechanisms involved in fruit ripening and the implementation of these regulatory modules as a promising breeding tool, with the potential of opening new avenues for the development of stress-tolerant, nutritious, high-yielding crops.
Connect with Javier: Twitter
 Vineeta Rai, Ph.D.
Vineeta Rai, Ph.D.
Postdoc in Entomology & Plant Pathology
Uncovering how plants shape the bacteria composition in their environment to improve growth and fitness
Research Blurb:
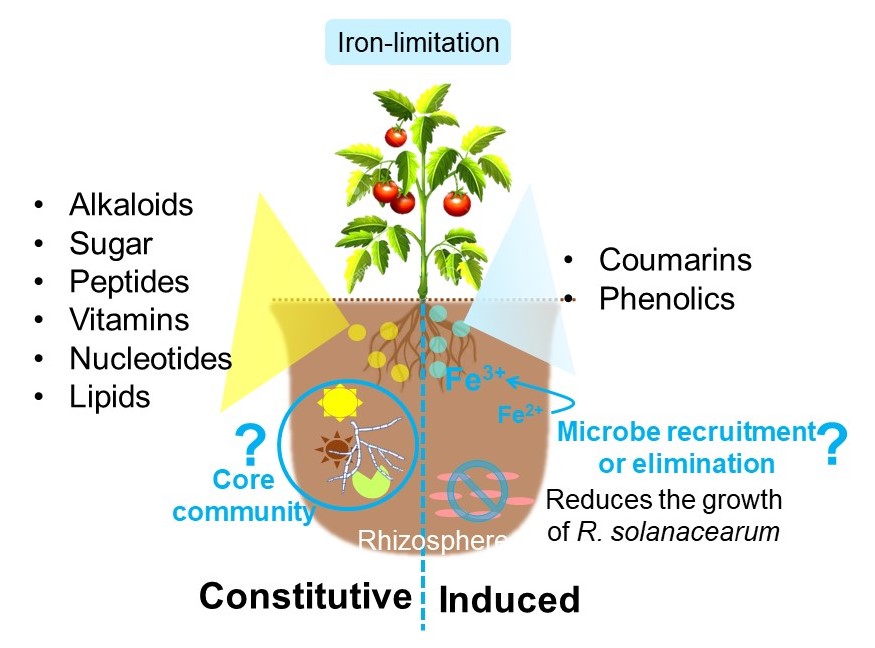
Plants thrive in virtually every environmental stress: abiotic and biotic. Plant roots are vulnerable as they directly face the various soil chemistry and soil microbiota. Roots exudate (secrete) 20–40% of the photosynthetically fixed carbon to establish rhizosphere biochemical networks comprised of low- or high-molecular-weight compounds. Low-molecular-weight compounds, including primary and secondary metabolites, are highly diverse. Root exometabolites can either be secreted constitutively or induced by biotic stress and abiotic factors like iron-limitation. Studies on the iron-limitation induced root exudation profiles of the model plant Arabidopsis showed coumarins as the major exometabolites. Coumarins are involved in iron solubilization and microbiome shaping. Interestingly, the root exudation profiles vary among plant species. So we are studying tomato root exudates to determine the common and unique metabolites between tomato and Arabidopsis. We chose tomato as the iron acquisition metabolism is similar to Arabidopsis (both are Strategy I plants); however, it still differs in the productions of other metabolites like alkaloids. We identified ~200 tomato root exometabolites using untargeted metabolomics. Iron-limitation induced the exudation of coumarins and phenolic compounds like N-trans-p-coumaroyloctopamine and feruloyltyramine in tomato. Iron-limitation induced metabolites viz. coumarins (coumarin, umbelliferone), and phenolic compounds (cinnamic acid) reduced the growth of the tomato bacterial wilt pathogen R. solanacearum. We are currently in the process of analyzing the growth of R. solanacearum against tomato root exudates. Decoding the root exometabolite composition is the first step towards the understanding of how plants shape the rhizosphere microbiome and soil chemistry. In the future, the findings may lead to the identification of specific mechanisms to combat low-nutrient supply and pathogens and improve plant fitness under non-ideal growth conditions in association with beneficial microbes.
Key Finding:
We identified ~200 root-secreted metabolites in tomato (Heinz 1706, genetically sequenced) by available mass spectrometry data-mining and assisted by plant genomic information. The major exometabolites profiled in two tomato varieties (Heinz 1706 and Hawaii 7996, Ralstonia solanacearum resistant) were similar in replete conditions, while the levels of aesculetin, a coumarin, differed in iron-limitation. The majority of primary metabolites within chemical classes: alkaloids, sugars, peptides, mono- and dicarboxylic acids, vitamins, nucleotides, and lipids were constitutively secreted, while iron-limitation induced the exudation of coumarins and phenolic compounds like N-trans-p-coumaroyloctopamine and feruloyltyramine. Iron-limitation induced metabolites viz. coumarins (coumarin, umbelliferone), and phenolic compounds (cinnamic acid) reduced the growth of the tomato bacterial wilt pathogen R. solanacearum.
Research Impact:
Pinpointing the constitutive and induced tomato root exometabolites in iron-limitation will lead to a better understanding of the biochemical networks mediating plant-microbe interactions, particularly in abiotic stress conditions.
Connect with Vineeta: LinkedIn
 Brenna Zimmer, Ph.D.
Brenna Zimmer, Ph.D.
Postdoc in Molecular and Structural Biochemistry
Identifying key biochemical markers associated with prostate cancer progression
Research Blurb:
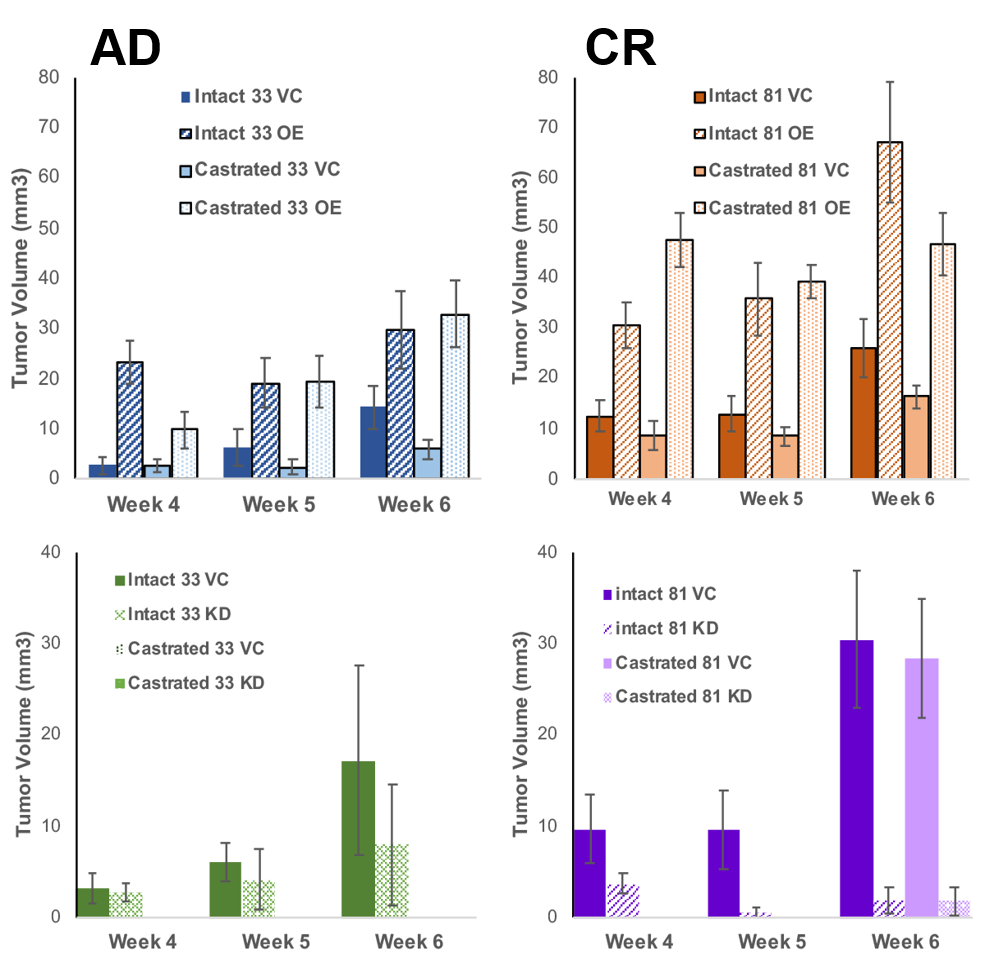
Prostate cancer is a devastating disease and is a leading cause of cancer-related death in Western men. Loss of androgen sensitivity following androgen deprivation therapy remains a clinical problem and leads to disease recurrence termed castration-resistant prostate cancer (CRPC). Understanding the underlying mechanisms that contribute to CRPC formation can help in a targeted therapeutic approach to treat disease and improve quality of life. My research is focused on elucidating these mechanisms to better our understanding on the biochemical changes that occur as disease progresses. Our lab has identified UDP-glucose dehydrogenase (UGDH) as a potential novel field-specific biomarker and have shown UGDH expression to clinically correlate with aspects of disease progression. Our work has shown that knocking down expression of UGDH leads cells to retain androgen sensitivity in culture. Conversely, overexpression of UGDH mimics the phenotype of CRPC. Importantly, manipulating UGDH expression altered the tumorigenic potential in mice. Overall, our results functionally implicate UGDH as a novel sensory node in the prioritization of prostate androgen inactivation and illustrates potential diagnostic and prognostic value of UGDH as a quantitative biomarker.
Research Impact:
Approximately 1 out of every 36 men develop this aggressive and lethal form of the disease. There is a less than 5-year life expectancy following diagnosis and treatment options are debilitating and costly. My hope is that our work will enhance point of care therapy for patients. By better understanding the disease, clinicians can make informed decisions for the best therapeutic approach for each patient and improve overall quality of life.
Connect with Brenna: LinkedIn
- Categories: